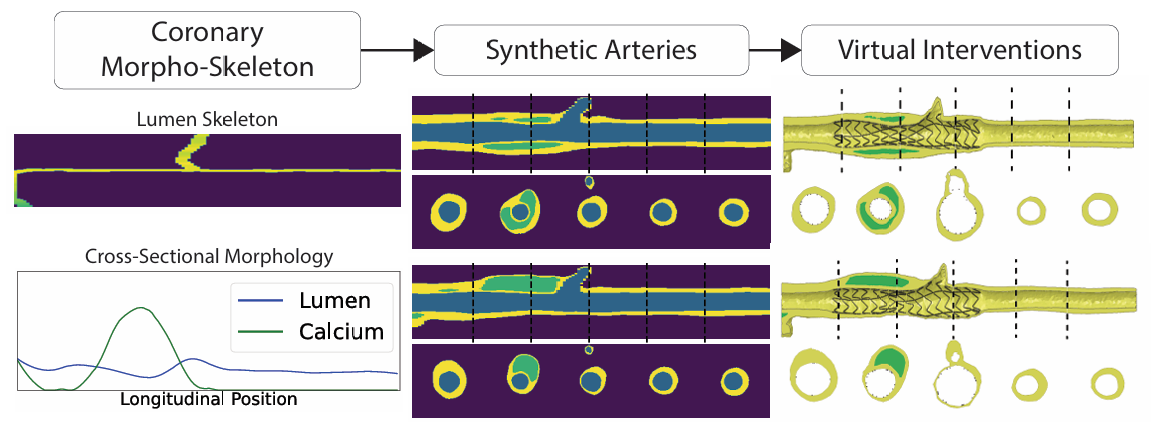
A Diffusion Model for Simulation Ready Coronary Anatomy with Morpho-skeletal Control
arXiv preprint, 2024. Karim Kadry, Shreya Gupta, Jonas Sogbadji, Michiel Schaap, Kersten Petersen, Takuya Mizukami, Carlos Collet, Farhad R Nezami, Elazer R Edelman
We leverage Latent Diffusion Models (LDMs) to generate realistic coronary artery anatomies for virtual intervention studies. Our approach balances controllability and realism, enabling the creation of anatomies based on topological, morphological, and skeletal constraints. This framework allows for device designers to derive mechanistic insights regarding anatomic variation and simulated device deployment.
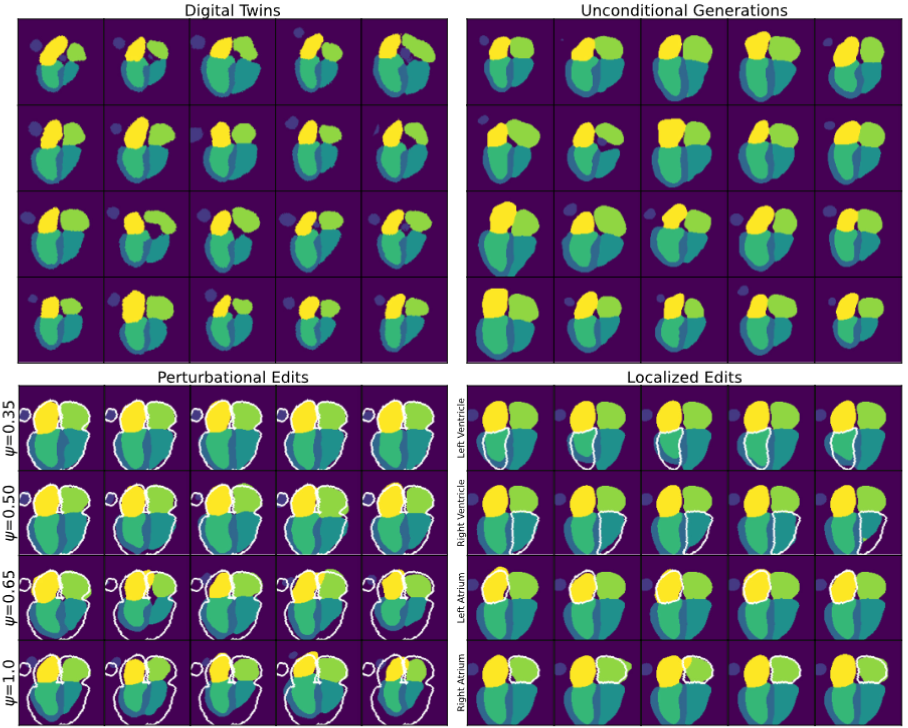
Probing the Limits and Capabilities of Diffusion Models for the Anatomic Editing of Digital Twins
arXiv preprint, 2023. Karim Kadry, Shreya Gupta, Farhad R Nezami, Elazer R Edelman
We explore the use of Latent Diffusion Models (LDMs) to edit digital twins of cardiac anatomy, creating anatomic variants termed digital siblings. These digital siblings enable comparative simulations to study the impact of subtle anatomic variations on cardiovascular device deployment. Our framework demonstrates the potential of LDMs for virtual cohort augmentation and addresses issues related to dataset imbalance and diversity.
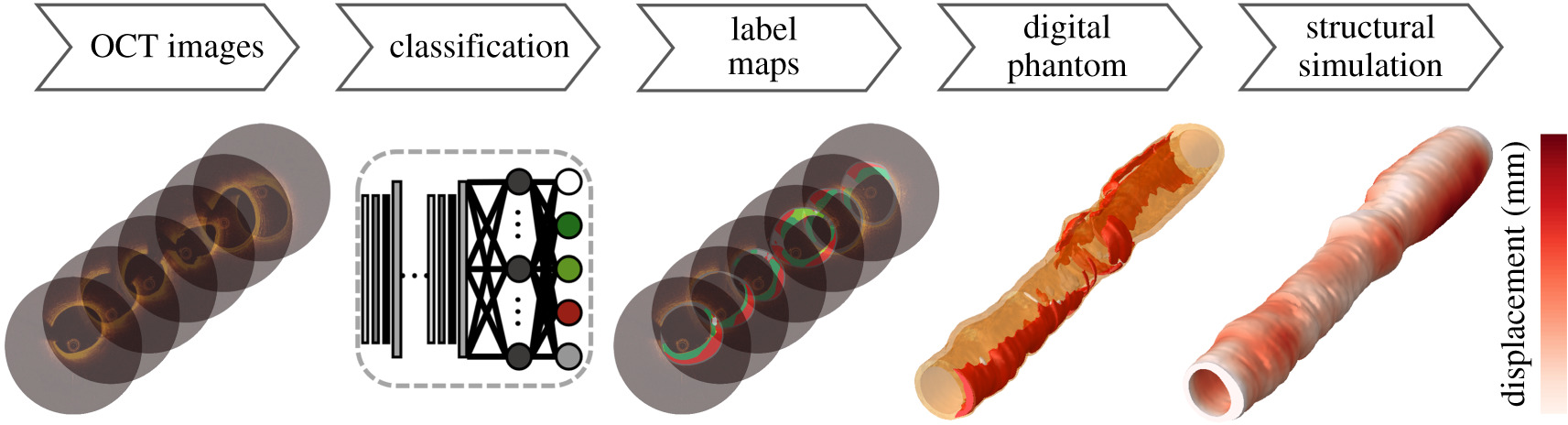
Royal Society Interface, 2021. Karim Kadry, Max L. Olender, David Marlevi, Elazer R. Edelman, Farhad R. Nezami
We develop an end-to-end computational pipeline for patient-specific coronary artery biomechanics from intravascular imaging and study the influence of various diseased tissues on mechanical wall stress.
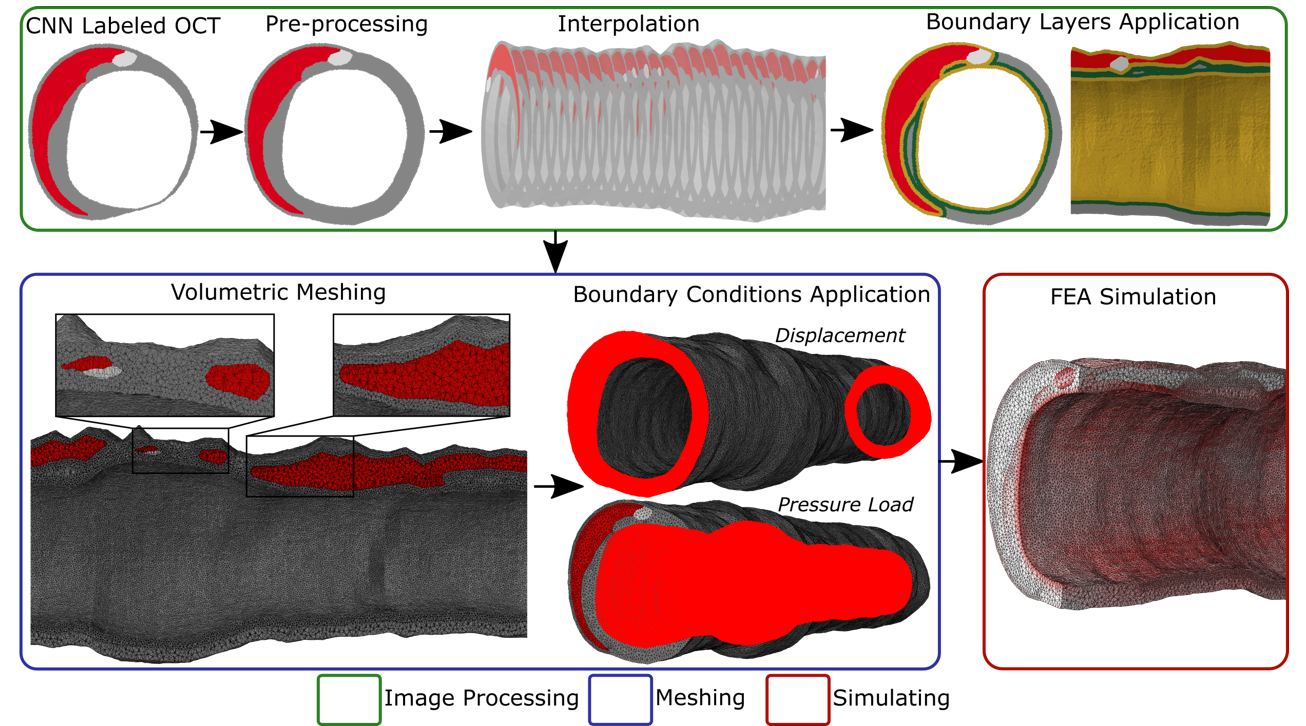
Computers in Biology and Medicine, 2023. Ross Straughan*, Karim Kadry*, Sahil A Parikh, Elazer R. Edelman, Farhad R. Nezami
We automate our previous computational pipeline for patient-specific coronary biomechanics, enabling the scalable execution of high-fidelity biomechanical simulations from large datasets of OCT images.
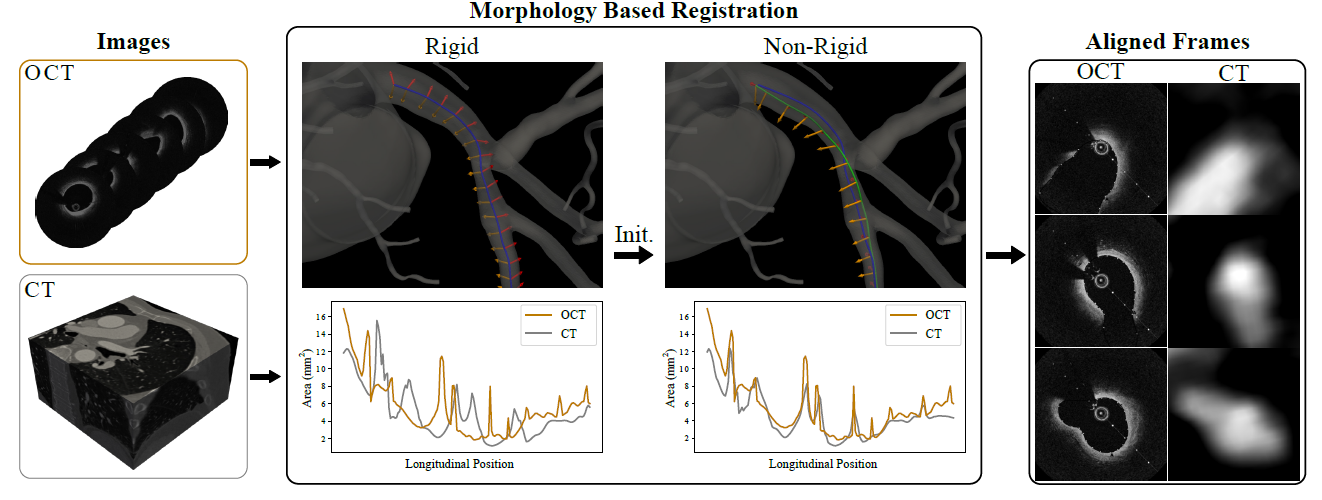
Arxiv, 2022. Karim Kadry, Abhishek Karmakar, Andreas Schuh, Kersten Peterson, Michiel Schaap, David Marlevi, Charles Taylor, Elazer Edelman, Farhad Nezami
We present a morphology-based framework for aligning intravascular and computed tomography (CT) images. This approach combines the high-resolution cross-sectional detail of intravascular imaging with the accurate 3D information from CT scans, enabling the creation of high-fidelity digital twins of coronary arteries.
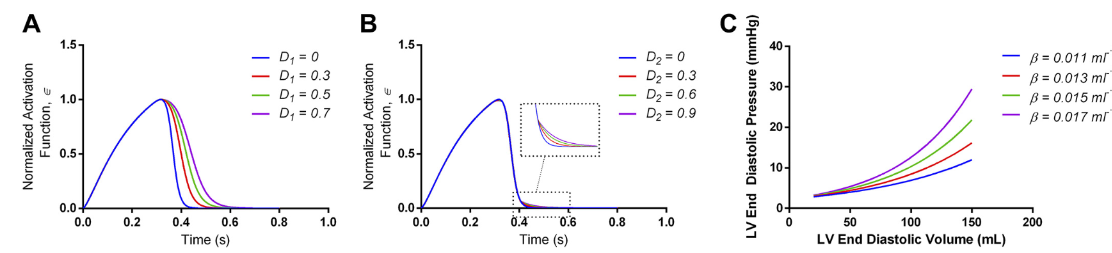
Biomechanics of Diastolic Dysfunction: A One-Dimensional Computational Modeling Approach
American Journal of Physiology-Heart and Circulatory Physiology, 2020. Karim Kadry, Stamatia Pagoulatou, Quentin Mercier, Georgios Rovas, Vasiliki Bikia, Hajo Müller, Dionysios Adamopoulos, Nikolaos Stergiopulos
We develop a computational model to study diastolic dysfunction (DD), a key component of heart failure with preserved ejection fraction (HFpEF). Our model integrates a one-dimensional arterial network with a zero-dimensional cardiac model to investigate the active relaxation and passive stiffness of the left ventricular wall. This framework provides insights into the biomechanical mechanisms of DD and HFpEF.
–>
